Recoding (Variable) Values in R
** Download the R syntax file and the data files referenced in the post **
Earlier this month, in another post, I introduced my "Codebook Method" along with my favorite options for quickly renaming variables in R. This post shares my favorite R recoding tips.
The first set of tips are for folks who want to recode values manually. The second part of this post shows (one way) how to automate value recoding tasks in R.
--
Like my last post, I generated a fake data set
and created a sample codebook. The codebook has five columns:
var_name: The original name assigned to each variable in the data set
variable.label: A description of the variable
type: Identifies what level of measurement the variable has
values: List of the values each variable can have
value.labels: Descriptions for each unique variable value
(Note: I omitted the "new_var" column as I am not renaming the variables in the data set.)
VALUE RECODING
Most people opt to use a programming language like R to automate data work. That said, all tasks have a manual equivalent. And while it may seem helpful to leapfrog to automated solutions, learning the (manual) basics gives you a springboard for developing creative and thoughtful solutions.
When recoding (variable) values, it is important to have three pieces of information:
Names of the variables to recode
Original values (usually numeric in nature)
New values (sometimes they are numeric but oftentimes they are string labels)
For this post, I focus exclusively on how to convert numeric values into string labels. Though the same tips can be applied to processes that require converting string labels to numeric values.
Setup
First, load the packages and import the fake data set into your R environment.
#### RECODING VARIABLE VALUES ####
## PACKAGES ##
# create notin operator
`%notin%` <- Negate(`%in%`)
# Download packages if not available
pckgs <- c("tidyverse", "data.table", "rlang", "readxl")
# load packages
sapply(pckgs, FUN = require, character.only = TRUE)
## Load Data Set ##
dat <- read_excel("recode_vals.xlsx", sheet = "fake_dat")
The data set has four variables:
ID: An identification variable
Gender: A variable that asked individuals to select from a list of options which best described their gender identity
Low Income: A variable indicating whether the individual's household is low income
Education Level: A variable indicating the highest level of education the individual has received
Part I: Manual Recoding
Option 1: Classic ifelse (Base R)/ if_else (dplyr)
When it comes to manually recoding variable values in R, everyone learns if/else statements. The ifelse function from Base R is pretty simple: if something then do something. You can even "nest" these statements to specify multiple conditions. The if_else function from dplyr works pretty much the same way as ifelse, just with one added restriction: the values supplied for the TRUE and FALSE conditions must be the same type.
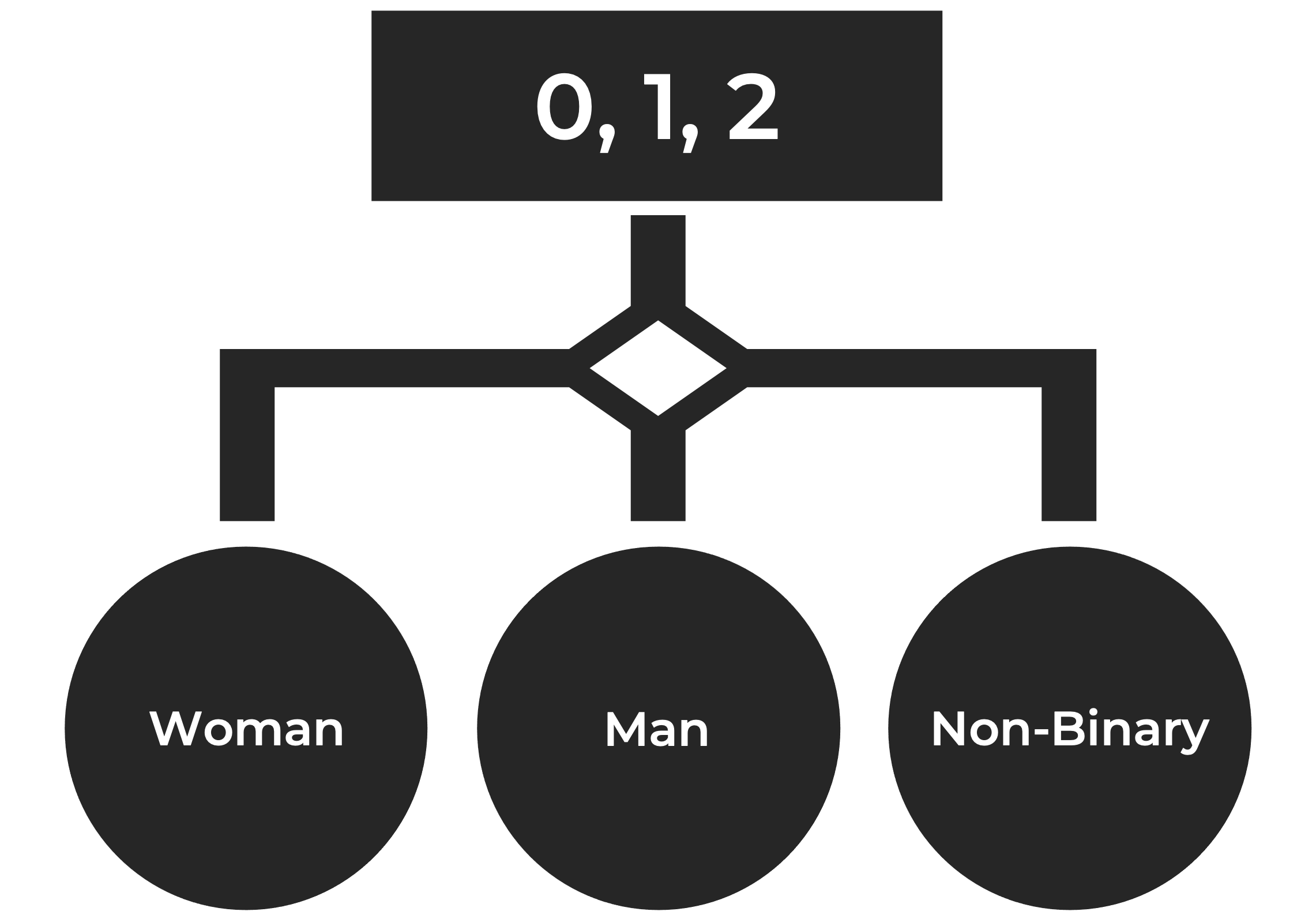
Let's say you are interested in recoding our variable "Gender" from the data set. In other words, you want to convert the 0s,1s, and 2s into labels (e.g., Woman, Man, Non-Binary). ifelse can easily accomplish this task:
## Option 1: if_else (base)
dat %>%
mutate(Gender = ifelse(Gender == 0, "woman",
ifelse(Gender == 1, "man",
ifelse(Gender == 2, "non-binary",
NA_character_))))
## Could also be written as ifelse (dplyr)
dat %>%
mutate(Gender = if_else(Gender == 0, "woman",
if_else(Gender == 1, "man",
if_else(Gender == 2, "non-binary",
NA_character_))))Note: A series of nested ifelse were used to recode the values.
Clearly, the downside of using this method is that if you have a nominal variable with many values, your code is going to be exceptionally long.
Option 2: case_when
(Packages used: dplyr)
Another option is the case_when function from the dplyr package. The case_when function works similarly to ifelse. However, where they differ is that case_when evaluates a condition using a two-sided formula: on the left-hand side is the logical test/condition and on the right-hand side is the value you wish to assign if that condition is TRUE. If you are evaluating multiple conditions, it is a good idea to also specify a *final* "TRUE" condition, which assigns a value or label to all other values that do not match the previously defined conditions.
## Option 2: case_when (dplyr)
dat %>%
mutate(`Low Income` = case_when(`Low Income` == 0 ~ "not low-income",
`Low Income`== 1 ~ "low-income",
TRUE ~ NA_character_))
Like Option 1, the case_when method can become laborious to write, especially if you need to convert a nominal variable with many values.
Option 3: recode + set_names
(Packages used: dplyr and rlang; both can be loaded from the tidyverse)
Yet another popular option is the recode function from the dplyr package coupled with the set_names function from the rlang package. recode, as its name suggests, replaces values in a vector based on their position or their name. set_names, on the other hand, creates a named vector. So when you combine recode with the power of set_names, you can explicitly set the vector values to be replaced as well as the vector of new labels to be applied. The best part? You can set both default and missing values.
## Option 3: recode + set_names (dplyr and rlang)
dat %>%
mutate(Gender = recode(Gender, !!!(set_names(c("woman","man","non-binary"), 0:2)), .default = NA_character_))
Like Options 1 and 2, however, this method can also lead to very lengthy code blocks.
Option 4: recode + deframe
(Packages used: dplyr and tibble; both can be loaded from the tidyverse)
Option 4 is almost identical to Option 3. The deframe function, like set_names,
creates a named vector, *BUT* it does so from a base two-column data frame. So, to use this method you must construct a two-column data frame that contains values and labels to replace the values.
## Option 4: recode + deframe (dplyr and tibble)
# Setup
key_edu <- tibble(values = 0:5, labels = c("Less than High School", "Some High School","High School Graduate", "Some College / Associate's Degree", "Bachelor's Degree", "Graduate Degree"))
dat %>%
mutate(`Education Level` = recode(`Education Level`,!!!(deframe(key_edu))))
Part II: Recoding Automated
All the methods I shared are practical ways to recode variable values. But, if you are like me, you want to implement a procedure that requires little effort and can easily be applied across a range of situations. The best way to do this: create a user defined function.
First things first, import the codebook. Be sure to omit the row where the variable name is "ID" (as we do not want to recode the ID values) and only keep relevant columns (i.e., var_name, values, and value.labels).
# import the codebook
key <- read_excel("recode_vals.xlsx", sheet = "codebook") %>%
select(c("var_name"),contains("value")) %>%
filter(var_name %notin% "ID")
Using the head function, print the first few lines of the key (i.e., codebook) data frame to your console. Note how the values (and value labels) are separated by a comma delimiter. Thus, to use this information in a recoding procedure, we must remove the delimiter:
## Functions
## split and unlist character values (separated by commas) into a numeric vector with n elements
vulist <- function(x){suppressWarnings(as.numeric(unlist(strsplit(as.character(x), split = ",")))) }
## split and unlist string (separated by commas) into a character vector with n elements
vlulist <- function(x){suppressWarnings(unlist(strsplit(as.character(x), split = ",")))}
Note: there are two such functions: (1) one for (numeric) values and (2) another for string labels.
Next, combine these functions into a larger function that performs the recoding task (packages used: rlang, dplyr, tibble):
The function I created takes the name of a column (i.e., a variable name), subsets the codebook by the variable name, and then creates a two-column data frame containing the variable's (original) values and labels. In the final step, the variable's values are recoded.
## Option 5: recode values function (deframe)
# (rlang, tibble, and dplyr)
recode_the_vals <- function(x){
x_name <- quo_name(enquo(x))
key.sub <- key %>% filter(var_name %in% x_name)
label.keys <- tibble(values = vlulist(key.sub$values), labels = vlulist(key.sub$value.labels))
recode(x, !!!(deframe(label.keys)))
}
Now when you recode values, you can take advantage of the function you created. The best part? It only requires two lines of code:
# recode values
dat %>%
mutate(across(.cols = -c(ID), ~ recode_the_vals(.x)))
See, a codebook can make a world of difference when developing and implementing a data workflow.
If you have a favorite method for recoding variable values in R, share your code in the comments below.
Need help designing a data cleaning pipeline? Get in Touch!